Developing best practices for genotyping-by-sequencing analysis in the construction of linkage maps
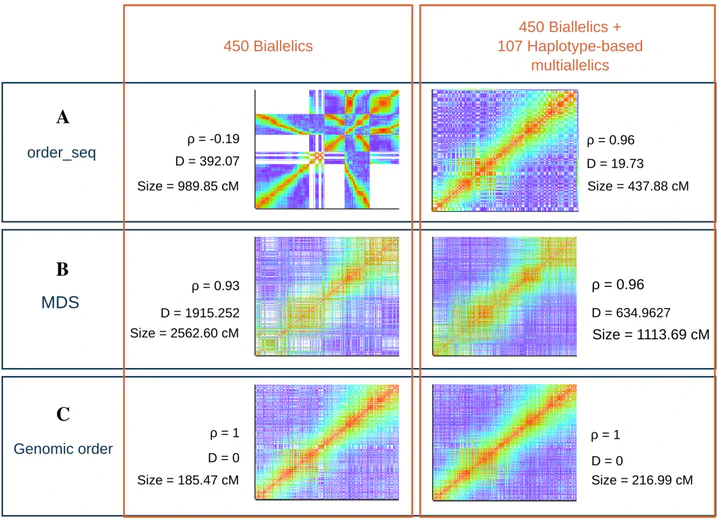
Genotyping-by-sequencing offers cost-effective, high-density marker data but also introduces PCR bias, sequencing errors, and contaminants that distort linkage maps. We benchmarked the Reads2Map workflows across simulated and empirical diploid outcrossing populations, pairing variant callers (GATK, Stacks, TASSEL, FreeBayes) with genotype callers (updog, polyRAD, SuperMASSA) and linkage-map tools (OneMap, GUSMap). We quantified when genotype-probability models, global error rates, or haplotype-based multiallelic markers best recover the expected genetic distances, and flagged scenarios where pipeline choices are dataset dependent. The resulting best-practice defaults, together with the Reads2MapApp visual diagnostics, reduce the trial-and-error burden when building reliable linkage maps from GBS data.